Load annotations from a CSV file
LoadAnnotationCSV.RdTakes a character vector with paths to CSV files and
returns a data.frame. The resulting data.frame
can be added to a Seurat object using the AddMetaData
function.
Details
NB: The files need to be loaded in the correct order. For example,
if you have an object with 5 tissue sections, you should provide
5 paths in the correct order.
See also
Other pre-process:
CreateMultiModalObject(),
LoadAndMergeMatrices(),
LoadImageInfo(),
LoadImages(),
LoadScaleFactors(),
LoadSpatialCoordinates(),
ReadVisiumData(),
UpdateImageInfo()
Examples
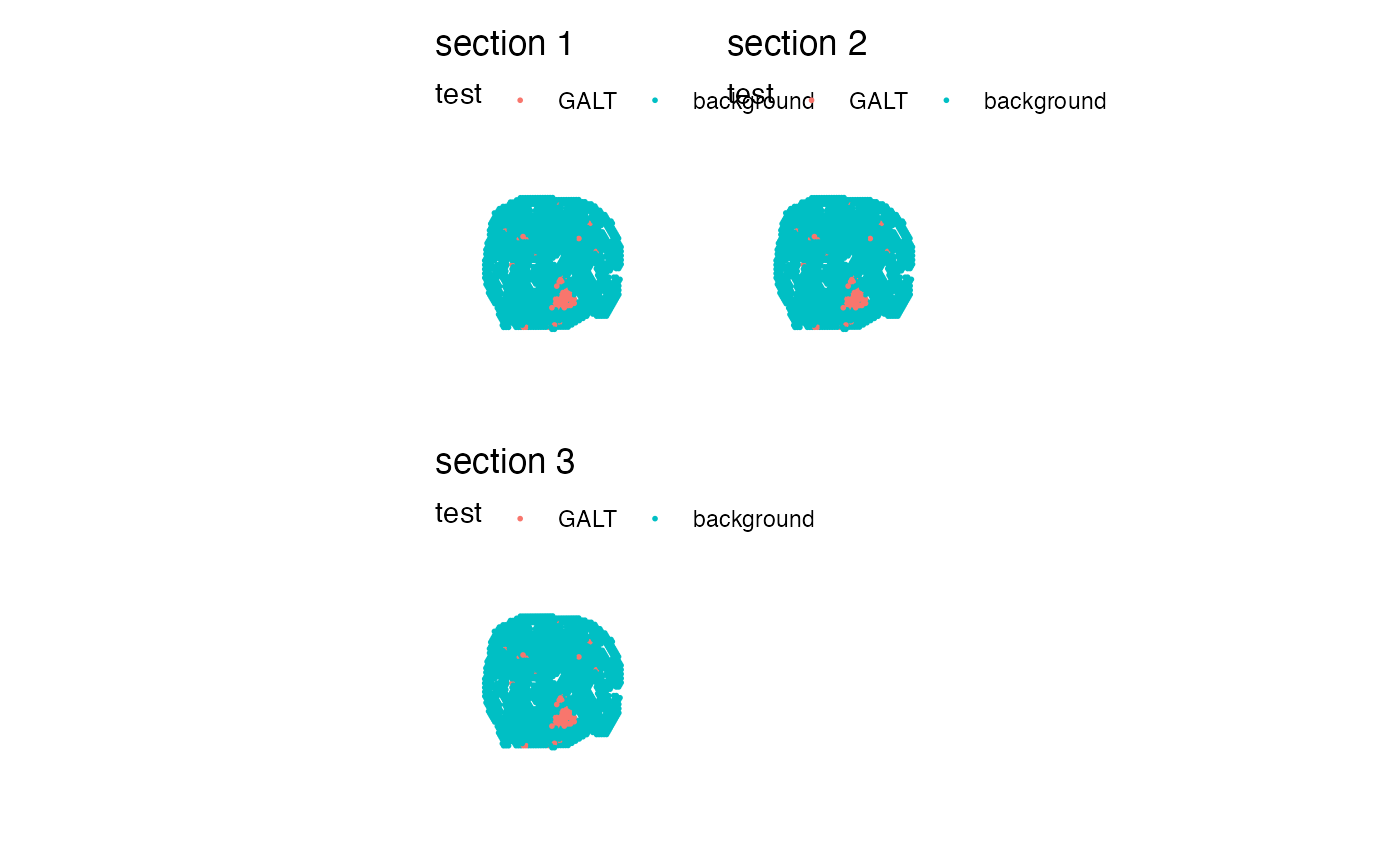
library(semla)
se_mcolon <- readRDS(system.file("extdata/mousecolon", "se_mcolon", package = "semla"))
se_merged <- MergeSTData(se_mcolon, y = list(se_mcolon, se_mcolon))
# Get annotation file(s)
annotation_files <- system.file("extdata/mousecolon",
"galt_spots.csv",
package = "semla") |> rep(3)
# Load annotations
annotations <- LoadAnnotationCSV(annotation_files)
head(annotations)
#> selection
#> AAACACCAATAACTGC-1 background
#> AAACAGAGCGACTCCT-1 background
#> AAACAGCTTTCAGAAG-1 background
#> AAACAGGGTCTATATT-1 background
#> AAACCGGGTAGGTACC-1 background
#> AAACCGTTCGTCCAGG-1 background
# Edit data.frame if needed
annotations <- annotations |>
rename(test = selection)
# Add annotations to meta data
se_merged <- AddMetaData(se_merged, metadata = annotations)
# Plot new categorical variable
MapLabels(se_merged, column_name = "test")