
Load images
load-images.RdLoad H&E images (or any other custom images) required for visualization methods in semla.
Usage
LoadImages(object, ...)
# Default S3 method
LoadImages(object, image_height = 400, pad_info = NULL, verbose = TRUE, ...)
# S3 method for class 'Seurat'
LoadImages(object, image_height = 400, verbose = TRUE, ...)default method
If a character vector of image paths are provided, the images will be loaded,
then down-scaled based on image_height and returned as a list of raster
objects. Only JPEG and PNG images are supported.
Seurat
If a Seurat object is provided, the images will be loaded as raster objects
and stored inside the Staffli object that is located in the tools
slot.
See also
Other pre-process:
CreateMultiModalObject(),
LoadAndMergeMatrices(),
LoadAnnotationCSV(),
LoadImageInfo(),
LoadScaleFactors(),
LoadSpatialCoordinates(),
ReadVisiumData(),
UpdateImageInfo()
Other pre-process:
CreateMultiModalObject(),
LoadAndMergeMatrices(),
LoadAnnotationCSV(),
LoadImageInfo(),
LoadScaleFactors(),
LoadSpatialCoordinates(),
ReadVisiumData(),
UpdateImageInfo()
Examples
library(semla)
# Get paths for example images
mousebrain_jpg <- system.file("extdata/mousebrain",
"spatial/tissue_lowres_image.jpg",
package = "semla")
mousecolon_jpg <- system.file("extdata/mousecolon",
"spatial/tissue_lowres_image.jpg",
package = "semla")
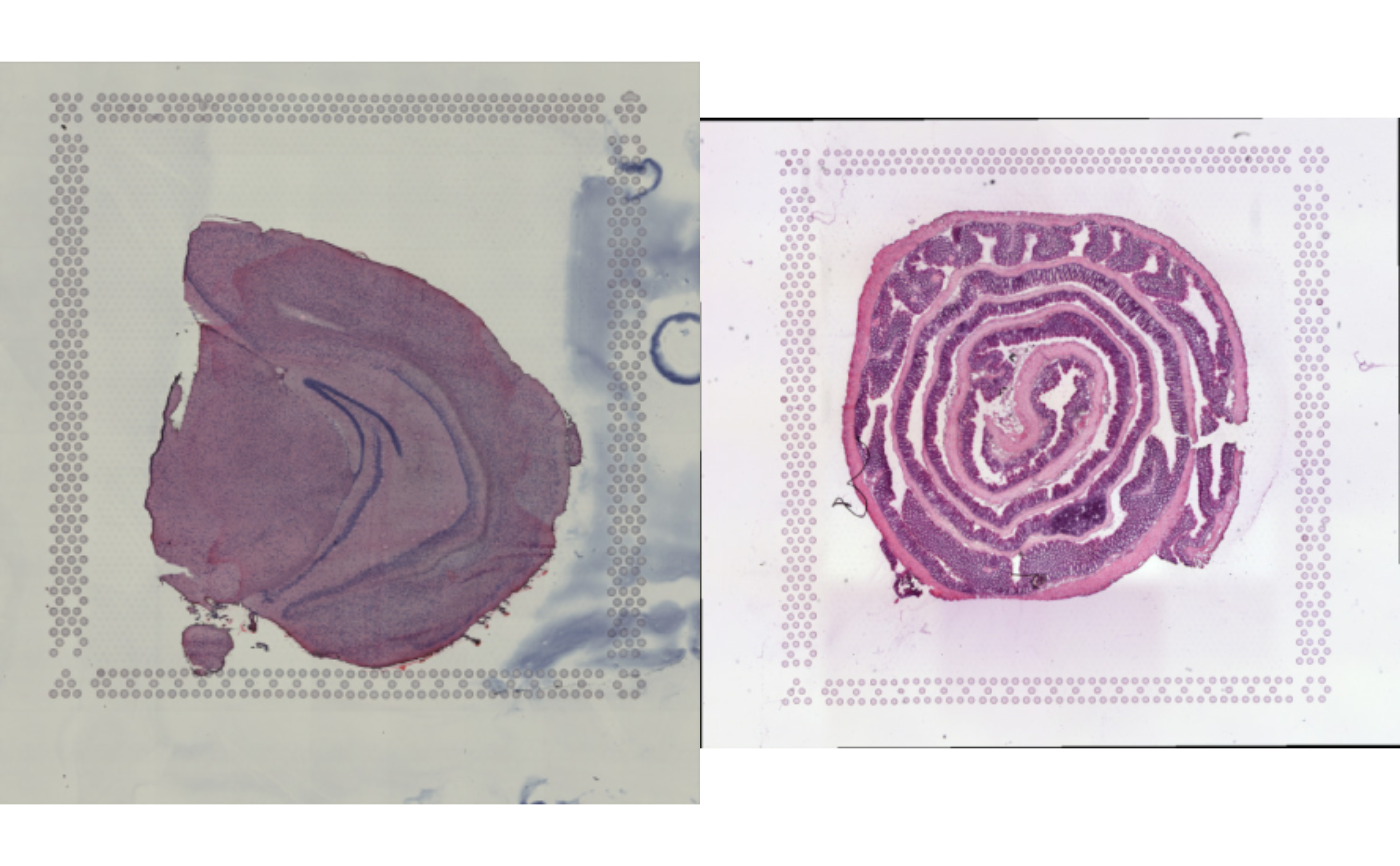
rasters <- LoadImages(c(mousebrain_jpg, mousecolon_jpg))
#>
#> ── Loading H&E images ──
#>
#> ℹ Loading image from /private/var/folders/z8/nrcst881607gn95xh3_4qvb00000gp/T/RtmpAdeL2K/temp_libpath109e41827971/semla/extdata/mousebrain/spatial/tissue_lowres_image.jpg
#> ℹ Scaled image from 600x565 to 400x377 pixels
#> ℹ Loading image from /private/var/folders/z8/nrcst881607gn95xh3_4qvb00000gp/T/RtmpAdeL2K/temp_libpath109e41827971/semla/extdata/mousecolon/spatial/tissue_lowres_image.jpg
#> ℹ Scaled image from 541x600 to 400x444 pixels
#> ℹ Saving loaded H&E images as 'rasters' in Seurat object
# Save graphical parameters
oldpar <- par(no.readonly = TRUE)
# plot images
par(mfrow = c(1, 2), mar = c(0, 0, 0, 0))
for (rst in rasters) {
plot(rst)
}
 # Reset graphical parameters
par(oldpar)
library(semla)
# Load example Visium data
se_mbrain <- readRDS(system.file("extdata/mousebrain", "se_mbrain", package = "semla"))
se_mcolon <- readRDS(system.file("extdata/mousecolon", "se_mcolon", package = "semla"))
se_merged <- MergeSTData(se_mbrain, se_mcolon)
# Load images
se_merged <- LoadImages(se_merged)
#>
#> ── Loading H&E images ──
#>
#> ℹ Loading image from /private/var/folders/z8/nrcst881607gn95xh3_4qvb00000gp/T/RtmpAdeL2K/temp_libpath109e41827971/semla/extdata/mousebrain/spatial/tissue_lowres_image.jpg
#> ℹ Scaled image from 600x565 to 400x377 pixels
#> ℹ Loading image from /private/var/folders/z8/nrcst881607gn95xh3_4qvb00000gp/T/RtmpAdeL2K/temp_libpath109e41827971/semla/extdata/mousecolon/spatial/tissue_lowres_image.jpg
#> ℹ Scaled image from 541x600 to 400x444 pixels
#> ℹ Saving loaded H&E images as 'rasters' in Seurat object
# Reset graphical parameters
par(oldpar)
library(semla)
# Load example Visium data
se_mbrain <- readRDS(system.file("extdata/mousebrain", "se_mbrain", package = "semla"))
se_mcolon <- readRDS(system.file("extdata/mousecolon", "se_mcolon", package = "semla"))
se_merged <- MergeSTData(se_mbrain, se_mcolon)
# Load images
se_merged <- LoadImages(se_merged)
#>
#> ── Loading H&E images ──
#>
#> ℹ Loading image from /private/var/folders/z8/nrcst881607gn95xh3_4qvb00000gp/T/RtmpAdeL2K/temp_libpath109e41827971/semla/extdata/mousebrain/spatial/tissue_lowres_image.jpg
#> ℹ Scaled image from 600x565 to 400x377 pixels
#> ℹ Loading image from /private/var/folders/z8/nrcst881607gn95xh3_4qvb00000gp/T/RtmpAdeL2K/temp_libpath109e41827971/semla/extdata/mousecolon/spatial/tissue_lowres_image.jpg
#> ℹ Scaled image from 541x600 to 400x444 pixels
#> ℹ Saving loaded H&E images as 'rasters' in Seurat object